Tutorials
This section contains a variety of tutorials that should help get you started with the oggmap package.
Getting started
If you are running oggmap for the first time, we recommend to either follow the individual oggmap steps or getting started with either the zebrafish case study (using OrthoFinder results) or the nematode case study (using pre-calculated orthomaps), which both cover all essential steps.
Pre-calculated orthomaps
In addition to extract gene age classes from OrthoFinder results, oggmap has the functionality to parse and extract gene age classes from pre-calculated orthologous group databases, like eggNOG or plaza.
If your query species is part of one of these databases, it might be sufficient to use the gene age classes directly from them and not start the time consuming step of orthologous group detection with OrthoFinder or any other related tool (see benchmark of tools at Quest for Orthologs).
Note
Since gene age class assignment for any query species relies on taxonomic sampling to cover at best all possible species tree nodes from the root (origin of life) up to the query species, the pre-calculated orthologous group databases might lack species information for certain tree nodes. Orthologous group detection algorithm do not account for missing species and as such will influence the taxonomic completeness score.
Note
To link gene age classes and expression data one should use the same genome annotation version for both, the orthologous group detection and the gene expression counting. To use the same genome annotation has the benefit not to miss any gene in one or the other and decreases the source of error during gene ID mapping.
eggNOG database version 6.0 orthomaps
includes 1322 species
Extracted orthomaps for all Eukaryota from eggNOG database version 6.0 can be downloaded here:
eggnog6_eukaryota_orthomaps.tsv.zip
To get an orthomap for e.g. the species Caenorhabditis elegans (taxID: 6239):
from oggmap import qlin, gtf2t2g, of2orthomap, orthomap2tei, datasets import pandas as pd eggnog6_eukaryota_orthomaps = pd.read_csv('eggnog6_eukaryota_orthomaps.tsv.zip', delimiter='\t') query_lineage = qlin.get_qlin(q='Caenorhabditis elegans') query_orthomap = eggnog6_eukaryota_orthomaps[eggnog6_eukaryota_orthomaps['taxID']==query_lineage[1]] query_orthomap
plaza database version 5.0 orthomaps
The plaza database offers two different sets of gene family clusters, either homologous (HOMFAM) or orthologous gene families (ORTHOFAM).
plaza dicots database version 5.0
includes 98 species
Extracted orthomaps for all dicots (HOMFAM and ORTHOFAM) from plaza dicots database version 5.0 can be downloaded here:
plaza monocots database version 5.0
includes 52 species
Extracted orthomaps for all monocots (HOMFAM and ORTHOFAM) from plaza monocots database version 5.0 can be downloaded here:
plaza_v5_monocots_HOMFAM_orthomaps.tsv.zip
plaza_v5_monocots_ORTHOFAM_orthomaps.tsv.zip
To get an orthomap for e.g. the species Arabidopsis thaliana (taxID: 3702):
from oggmap import qlin, gtf2t2g, of2orthomap, orthomap2tei, datasets import pandas as pd plaza_v5_dicots_HOMFAM_orthomaps = pd.read_csv('plaza_v5_dicots_HOMFAM_orthomaps.tsv.zip', delimiter='\t') query_lineage = qlin.get_qlin(q='Arabidopsis thaliana') query_orthomap = plaza_v5_dicots_HOMFAM_orthomaps[plaza_v5_dicots_HOMFAM_orthomaps['taxID']==query_lineage[1]] query_orthomap
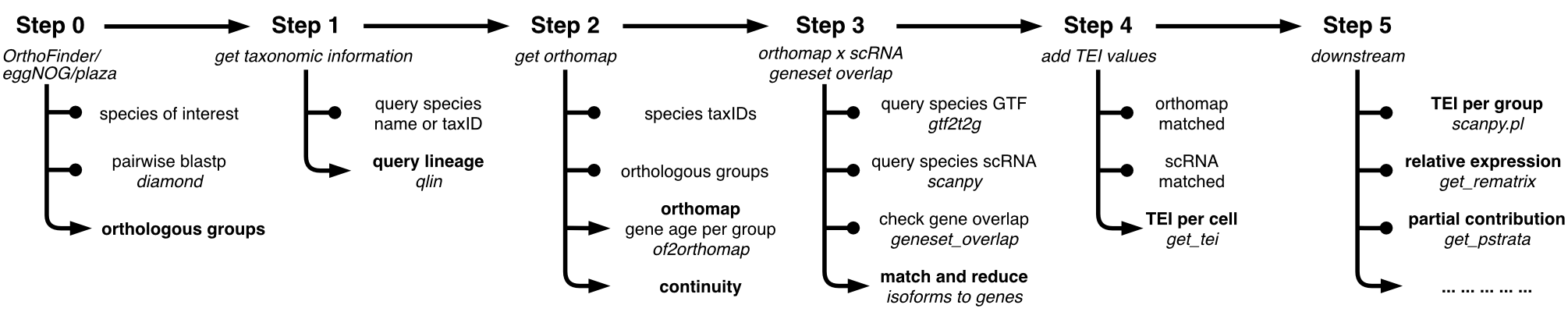
oggmap - Steps
This section contains the main steps of oggmap to extract gene age information for a query species up to linking the extracted gene age classes and expression data of single-cell data sets.
Step 0 - run OrthoFinder: This tutorial introduces how to run your own OrthoFinder analysis.
Step 1 - get taxonomic information: This tutorial introduces how to get taxonomic information.
Step 2 - gene age class assignment: This tutorial introduces how to extract an orthomap (gene age class) from OrthoFinder results or how to import pre-calculated orthomaps.
Step 3 - map gene/transcript IDs: This tutorial introduces how to match gene or transcript IDs between an orthomap and scRNA data.
Step 4 - TEI calculation: This tutorial introduces how to add a transcriptome evolutionary index (short: TEI) to scRNA data.
Step 4 - other evolutionary indices: This tutorial introduces how to use other evolutionary indices like nucleotide diversity to calculate TEI.
oggmap - Downstream analysis
This section contains different downstream analysis options (Step 5).
plotting results: This tutorial introduces some basic concepts of plotting results.
relative expression: This tutorial introduces relative expression per gene age class and its contribution to the global TEI per cell or cell type.
partial TEI values: This tutorial introduces partial TEI and its contribution to the global TEI per cell or cell type.
Case studies
hematopoiesis - case study: Notebook - Mus musculus hematopoiesis scRNA data example.
nematode embryogenesis - case study: Notebook - Caenorhabditis elegans embryogenesis scRNA data example.
zebrafish embryogenesis - case study: Notebook - Danio rerio embryogenesis scRNA data example.
frog embryogenesis - case study: Notebook - Xenopus tropicalis embryogenesis scRNA data example.
mouse - case study: Notebook - Mus musculus embryogenesis scRNA data example.
hydra - case study: Notebook - Hydra vulgaris cell atlas scRNA data example.
Note
A demo dataset is available for each of the tutorial notebooks above. These datasets allow you to begin exploring oggmap even if you do not have any data at any step in the analysis pipeline.
Command line functions
Command line functions: This section highlight all oggmap functions that can be run via the command line.
myTAI - Function correspondance
Correspondance of myTAI and oggmap functions: This tutorial covers which oggmap functions correspond to myTAI functions.
Prerequisites
This tutorial assumes that you have basic Python programming experience. In particular, we assume you are familiar with using a notebook from the following python data science libraries: jupyter.
To better understand plotting and data access, the user should try to get familiar with the python libraries: pandas, matplotlib and seaborn.
oggmap is a python package but part of it can be run on the command line. For the installation of oggmap, we recommend using Anaconda (see here). If you are not familiar with Anaconda or python environment management, please use our pre-built docker image.
Code and data availability
We provide links for the notebook in each tutorial section.
You can download the demo input data in the notebooks using the oggmap.datasets module.